As the number of new journal titles steadily increases from year to year, choosing the most appropriate publication outlet for a manuscript is becoming more complex than ever. Add to that the very real threat posed by predatory publishers who are actively trying to deceive unsuspecting authors – not to mention the multiple open access options available to choose from – and you have a journal selection process severely in need of some support.
Luckily, automation has come to the rescue! In this case, via the development of a category of journal selection/finder tools often referred to as Journal/Manuscript Matching Tools.
What are Journal/Manuscript Matching tools?
Journal/Manuscript Matchers are innovative tools that offer suggestions for which journals to consider submitting a manuscript to based on a text-matching search using the manuscript’s title and abstract provided by the author. This software is similar to plagiarism detection software in that it compares strings of text. However, the two tools likely apply very different cutoff values for variables like percent similarity. In the case of journal finders, they don’t run the text comparison against other full-text articles, but rather against bibliographic literature database records (ie. other titles and abstracts).
The Journal/Manuscript matching tools work under the assumption that journals that have published papers on a similar topic before may be more likely to be interested in publishing on this topic again in the future. By searching for published papers that are “similar” (as per text word match based on the title and abstract only) to an author’s manuscript, these tools are able to suggest potential journal homes where “similar” papers have been accepted before for publication. Generally, these tools are helpful in identifying appropriate journals in terms of scope and target audience.
A limitation, however, is that past journal editor behavior does not always predict future journal editor decisions. (For example, a journal that is broader in scope may have recently published a special issue on a particular topic or subject area that the editors are not planning on revisiting any time in the very near future.)
What are some examples of available Journal/Manuscript Matching Tools?
-
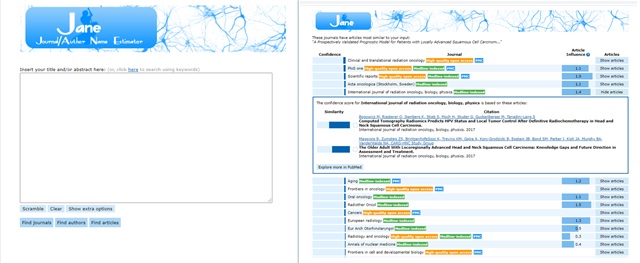
JANE: Journal/Author Name Estimator
-
Elsevier® Journal Finder
-
Clarivate Manuscript Matcher (Web of Science / EndNote)
-
Springer/Nature Journal Suggester
-
IEEE Publication Recommender
-
Taylor & Francis Journal Suggester
-
Wiley Journal Finder
-
JSTOR Text Analyzer
Which Journal/Manuscript Matcher should you use?
Depending on the discipline and subject area whose audience you are targeting, one of these options may be more fruitful/relevant – with regards to the suggestions made – than the others. For example, the JANE or Journal/Author Name estimator, conducts the text-matching search against the PubMed database and is best for targeting biomedical journal titles. As such, if you are targeting an engineering journal, for example, you may be better off exploring the suggestions provided by the IEEE publication recommender, or more multidisciplinary journal matchers like Elsevier’s or Clarivate’s journal finder tools.
The oldest of these tools, JANE, was developed in 2007 by a Biosemantics research group at the University Medical Center Rotterdam’s Department of Medical Informatics (Netherlands). It is the least biased towards a particular publisher’s journal titles and it offers a fairly current snapshot of publication trends as the data from PubMed that it runs the comparisons against is updated monthly.
To get a better understanding of what you get out of these journal selection tools in general and to learn more about JANE, see this recent review:
Curry CL. Journal/Author Name Estimator (JANE). J Med Libr Assoc. 2019 Jan;107(1):122–4. doi: 10.5195/jmla.2019.598. Epub 2019 Jan 1. PMCID: PMC6300233. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6300233/
To submit a request for assistance with the journal selection process, be sure to Ask Us at the MSK Library!



