PubMed – the National Library of Medicine’s database of biomedical literature that will celebrate its 30th anniversary in January 2026 – is an incredible resource that’s freely available to all.
So much so in fact that many other search tools, including Google Scholar and most of the generative AI research assistants that are popping up at a dizzying speed, heavily rely on PubMed for their content needs, especially since the National Center for Biotechnology Information (NCBI) has always been eager to build APIs and other tools to help facilitate collaborative relationships with developers of other research tools.
One of the downsides, however, of discovering PubMed’s content solely via other search engines and tools is that users miss out on some of the incredible value-added links to other information that appear within each PubMed record. This is particularly true for searches on topics with a genetic information or bioinformatics aspect.
Take this example (inspired by NLM training exercises):
You are interested in exploring how the CYP2r1 gene might impact vitamin D deficiency risk.
A basic search in PubMed might look something like this:
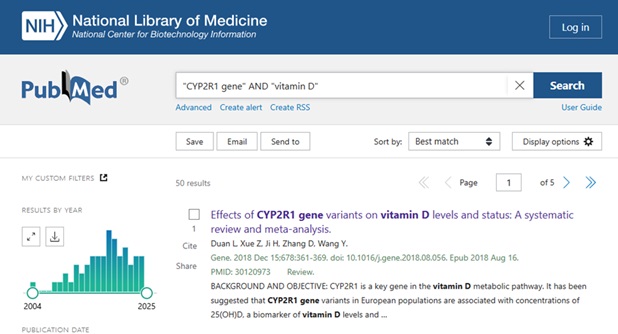
Clicking on the Title to view the Full Abstract view, users can scroll below the abstract text to see the MeSH terms and other Related Information – see:
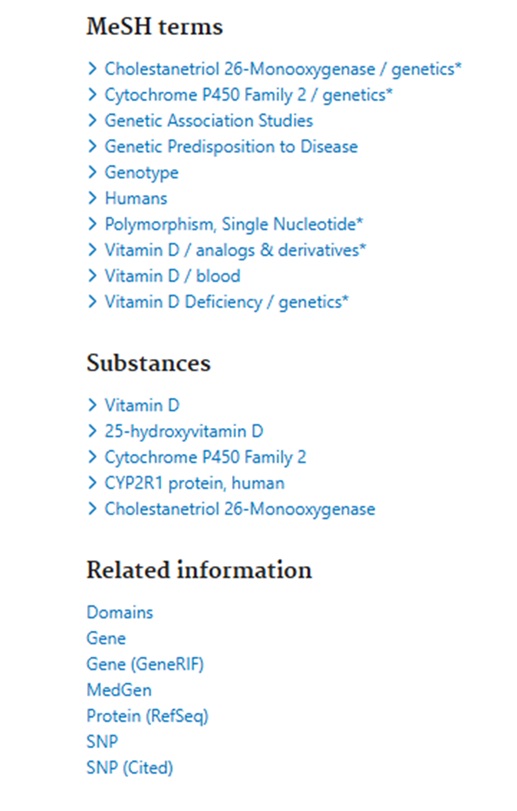
The Related Information links include a link to NCBI’s Gene information portal which:
“integrates information from a wide range of species. A record may include nomenclature, Reference Sequences (RefSeqs), maps, pathways, variations, phenotypes, and links to genome-, phenotype-, and locus-specific resources worldwide.”
The Gene record can also include GeneRIFs or a “Gene Reference into Function”.
See https://www.ncbi.nlm.nih.gov/gene/about-generif for a more detailed description.
“GeneRIF provides a simple mechanism to allow scientists to add to the functional annotation of genes described in Gene.”
As per https://www.ncbi.nlm.nih.gov/books/NBK3841/#EntrezGene.Bibliography:
“A GeneRIF is a concise phrase describing a function or functions of a gene, with the PubMed citation supporting that assertion.”
Filtering out the references that address a specific gene’s function can be a useful time-saver when literature searching.
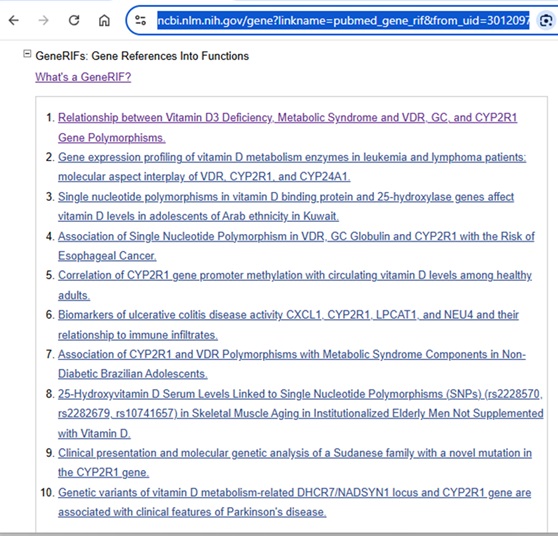
For those who find the Gene records a bit overwhelming and prefer to stay within the familiar PubMed environment, limiting PubMed search results to those items that have been added as Gene RIFs can be filtered out in a PubMed search by adding “pubmed gene rif” [Filter].
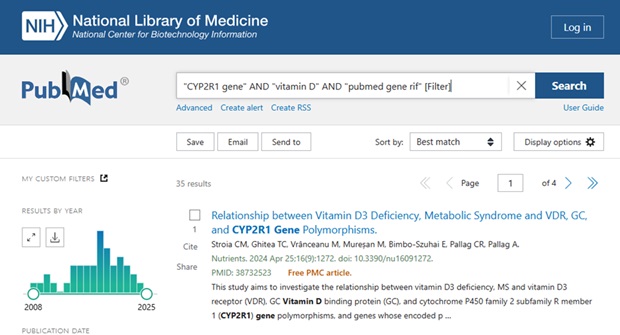
For example, adding it to the PubMed search string:
“CYP2R1 gene” AND “vitamin D” AND “pubmed gene rif” [Filter]
If you have any questions or want additional guidance on designing specialized literature searches, feel free to Ask Us at the MSK Library.