- Israeli scientists discovered that white blood cells, known as eosinophils, can help the body fight cancer, particularly cancer metastasis in the lungs. An original function of eosinophils is to aim the immune system at fighting parasites; they are also the cause of allergies.The findings from the animal study published in Cancer Research can lay a foundation for novel cancer treatments in the future.
- Researchers from Johns Hopkins Kimmel Cancer Center Convergence Institute conducted a retrospective analysis of large biomarkers’ datasets from tumors and healthy tissue. They found that older cancer patients could benefit as much as younger patients from cancer immunotherapies. The data suggested that biomarkers for immunotherapies could still be used as a basis to select immunotherapy treatment in older patients as in young ones, even though it is known that immunotherapy response weakens with aging. The study, which can potentially increase the use of immunotherapy in older population, was published in Cell Reports.
- A group of researchers from Ireland and France published a study on using microwave imaging to detect breast cancer. Potentially, this could be promising non-invasive technology with fewer side effects than currently used imaging modalities. The study was published in Academic Radiology.
- Swedish population-based study found a correlation between long-term (more than six months) use of antibiotics and risk of developing colon cancer five to ten years down the line. However, it is important to note that while epidemiological studies can establish a correlation, they can’t prove causation. The study was published in the Journal of the National Cancer Institute.
- Researchers from Japan conducted a study that established a correlation between the tumor size in poorly differentiated hepatocellular carcinoma (HCC) and the risk of early recurrence and patient survival, thus paving the way to improving the treatment of HCC patients. The study was published in Liver Cancer.
Author Archives: Marina
Pubmed Single Citation Matcher
Need to find a particular citation and don’t have the complete information about it?
Use a guided search with PubMed’s Single Citation Finder (the link is located on Pubmed main page in the Find category below the Search Box).
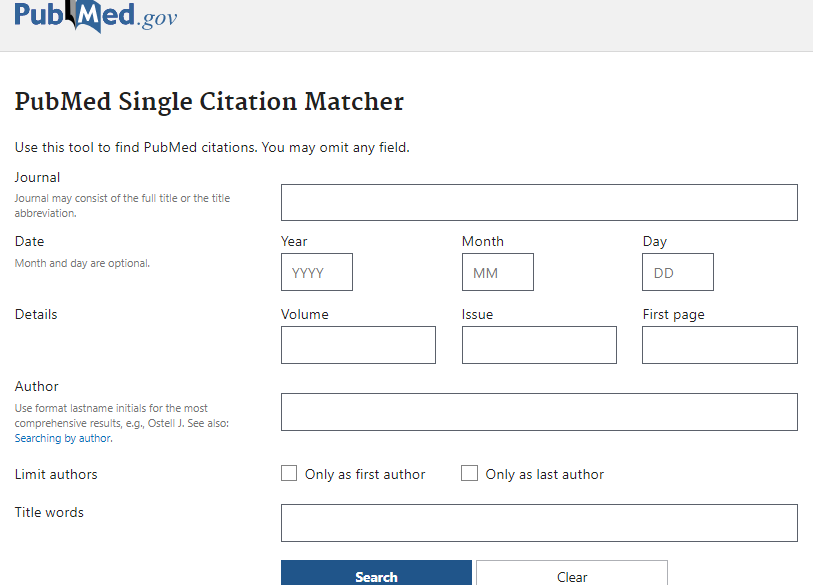
Single Citation Matcher guides you in entering information in pre-set search boxes dedicated to specific searchable fields in a Pubmed record, e.g. Journal, Title, Author.
If you don’t have all information about the article, enter only information you have at your disposal. The more information you enter the less search results you will get as your search will be more precise. Alternatively, if you enter very little information you will get more search results but you may still be able to get to the reference in question faster than by doing a general search in Pubmed.
You can use this tool for other purposes as well. For example, you can only use the Journal field to be able to browse the journal’s content, which you can do efficiently if your search results sorting order is Most Recent. To use the Journal field in Single Citation Matcher just start typing the name of the journal and then select the name, following the prompts.
Enjoy the convenience of this tool!
Embase: A Refresher
Embase, linked from the Library homepage under Top Databases, is a proprietary database, produced in Netherlands by Elsevier publishing company. It indexes journals in Medicine, Dentistry, Veterinary Science, Life Sciences, Public Health, Nursing, etc. While its coverage has significant overlap with PubMed (it actually indexes all of Medline), it also indexes a large number of international journals not found in PubMed. Embase also indexes supplements such as conference abstracts, clinical trials, and more.

Similar to PubMed’s MeSH terms, Embase also has the ability to map search terms to subject headings. Embase’s subject headings are called Emtree terms and their classification also has a hierarchical structure. One major difference between PubMed (MeSH) and Embase (Emtree) is that in PubMed narrower terms are automatically included, whereas in Embase, Emtree terms must be “exploded” to include all narrower terms found beneath a specific Emtree term.
Embase also includes some functions that are not found in PubMed, such as proximity searching, which besides AND, OR,and NOT, adds a layer to searches to make them more specific, using NEAR and NEXT.
Embase is typically one of the databases of choice used in searches when conducting a Systematic Review or a Meta-Analysis in biomedicine. It is recommended to specify the platform on which Embase was used. Embase is available on its native Elsevier platform (Embase.com) or on the OVID platform. MSKCC Library offers Embase on the Elsevier platform. The platform has an impact on the way the searches are conducted, so it is important to note when conducting systematic reviews.
Note: Starting July 1, 2021 Embase now requires signing into your Embase account to export citations to Endnote and other citation management tools. It is free to create an Embase/ Elsevier account and this login can be used for any Elsevier product (Embase, Scopus, etc.).