To make the most of the full text that’s available in PubMed Central (PMC), its search was recently updated!
For those new to this resource, PMC is a free full-text archive of biomedical and life sciences journal literature hosted by the National Library of Medicine (NLM). While PMC is a part of PubMed, and your searches in PubMed will undoubtedly show you some PMC results, not everything in PubMed will have full text immediately available to you. Starting your search directly in the PMC search interface means you’ll only get full-text PMC results.
PMC’s new search lets you harness that full text in new ways. To learn about everything new or updated, or how to use PMC, explore the PMC User Guide.
The new features I’m most excited about are:
- The ability to use proximity searching (also known as adjacency searching) within 14 different fields of a record. An example of this is “cancer pain”[ti:~1]. The double quotation marks and the information within the square brackets will pull in any record where cancer and pain are adjacent to each other by one or less terms in its title. It will bring in the phrases “cancer pain” as well as “pain of cancer.”
- Updated truncation searching that allows for unlimited variations of a term, instead of only the first 600 variations. An example of this is intervention*[tiab]. The asterisk and the information within the square brackets will pull in any record where a term that starts with intervention appears in its title or abstract. It will bring in the terms intervention, interventions, and interventional.
- The search field tag of [body] to search the full text of a record, meaning it searches all words and numbers in the body of an article, but not the abstract or references. An example of this is coping skill*[body]. The asterisk and the information within the square brackets will pull in any record where a phrase that starts with “coping skill” appears in its body. It will bring in the phrases “coping skill” as well as “coping skills.”
Below is an example of a search that showcases the new features I listed above, each connected with the Boolean AND. You can copy and paste this into the PMC search bar or just click on the hyperlink to be taken to a list of results.
“cancer pain”[ti:~1] AND intervention*[tiab] AND coping skill*[body]
While I’ve been enjoying the new PMC search, two things to note:
- A missing search feature requires your feedback.
- The federal government shutdown means you can’t give feedback.
There’s been some library listserv discussion about what was lost when PMC made the search switch, such as not being able to combine search strings or select a filter for language.
PMC Beta Search was introduced in April 2025 to give users a preview of the new search that went live in September 2025. Digging into recent PMC search announcements, there was subtle mention that some features would no longer be available once PMC Beta Search became the default. An August 2025 announcement built on an earlier April 2025 announcement and included the line “Some features of the current PMC search will not be included in this initial release.”
A notice on the PMC User Guide, included as a screenshot below, includes the line “the new PMC search represents a minimum-viable-product (MVP) approach.” The surrounding text implies a decision was made to deliver this new search experience with a focus on speed and not perfection, and that more search features could be coming based on user feedback.

Taken as a whole, you can see that anything you think is missing from PMC search needs your input in order to be known and prioritized. The two announcements and notice all recommend reaching out to the NLM Help Desk with feedback.
However, October 1 was the first day of the ongoing federal government shutdown. The NLM Help Desk and PMC are government-run, and consequently, no suggestions to improve PMC search can be received or implemented at this time.
But we MSK librarians are here! If at any time you have questions about PMC or searching in general, please reach out.
 In the example above, three requirements are worth highlighting:
In the example above, three requirements are worth highlighting: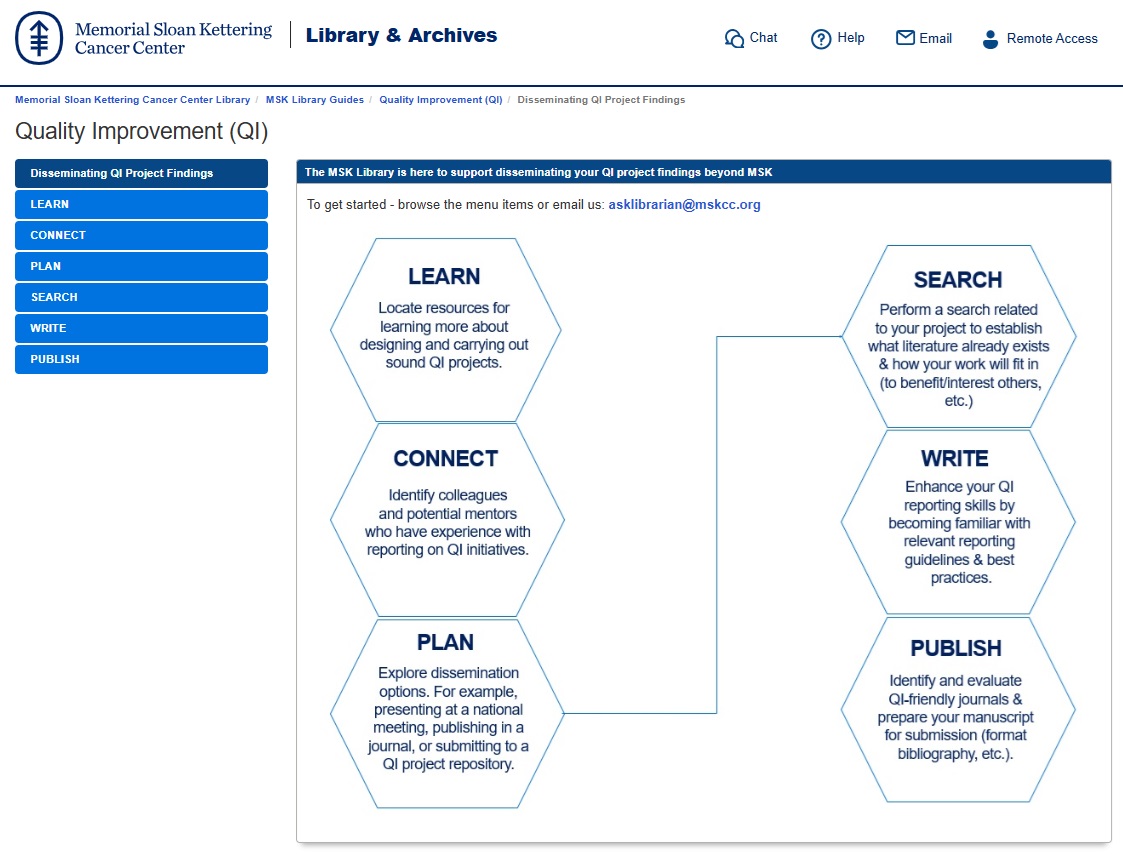 In particular, the WRITE module highlights resources helpful for becoming familiar with relevant reporting guidelines & best practices, while the LEARN module identifies resources for learning more about designing and carrying out sound QI projects.
In particular, the WRITE module highlights resources helpful for becoming familiar with relevant reporting guidelines & best practices, while the LEARN module identifies resources for learning more about designing and carrying out sound QI projects.