Cochrane Library is a collection of databases produced by the Cochrane Collaboration, that include Cochrane Systematic Reviews, Cochrane CENTRAL Register of Controlled Trials, and more.
Although Cochrane Library offers a MeSH search functionality, many searchers use just keyword searching so that not to duplicate the effort if they’ve already searched with MeSH in Pubmed or with Emtree terms in Embase (these 2 databases are major sources of Cochrane CENTRAL content).
There are four main ways to search Cochrane Library using keywords. The main difference between these four types of keyword searches is what is going on behind the scenes and how much control is given to the user in creating their search.
Basic Search
Cochrane’s most basic search is the search bar found on their homepage. It defaults to searching title, abstract, and keyword – however it gives little ability to modify searches or create comprehensive search strategies.
This search should only be used for the most basic and general searches, and not for conducting any type of literature review search.
The basic search also offers users to browse pre-set topics. These topics will automatically execute search on that topic to find Cochrane Reviews and Review Protocols, but it will not include results found in Trials.
Once a search is conducted using the basic search bar, the results are immediately displayed but also, the search string will appear within the advanced search page and the options below will become available for searches going forward.
Search Tab
The Search tab can be found by clicking on Advanced Search on the Cochrane homepage, just under the basic search bar; this takes you to the advanced search tabs. The main Search tab allows for more control of how and what is searched.
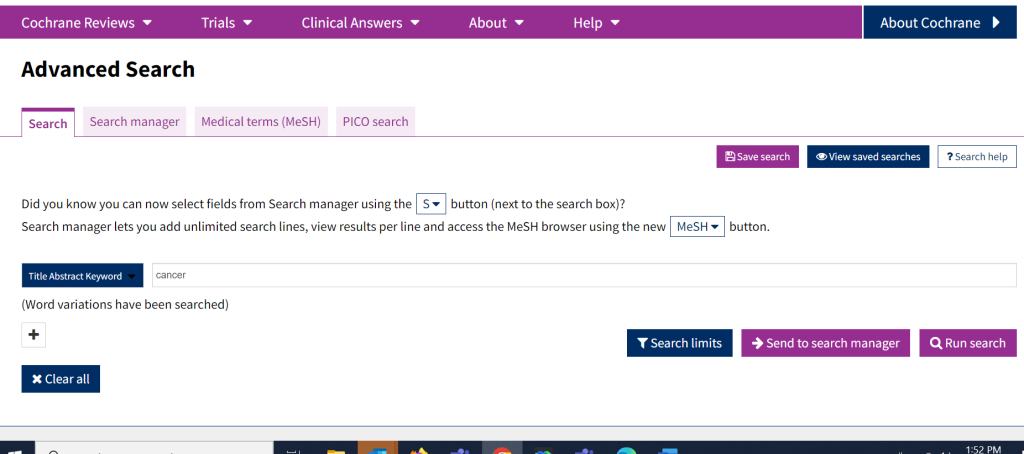
First, it allows for multiple search lines (that can be combined with Boolean operators), so you can combine multiple terms/concepts together easily. By clicking on the + it adds a new search box and from there you can identify what Boolean operator (AND, OR, NOT) you wish to use to combine the terms.
The advanced search tab also allows users to search a specific field (as opposed to just title/abstract/keyword). It also has search limit ability, to focus results by publication date or content type.
The one limitation of the search tab is it has limits on the length of search string used, similarly to basic search. The solution is to select Send to Search Manager instead of clicking Run Search.
Search Using Word Variations
One benefit of using the Search Tab (as compared to using the more functionally complex Search Manager), is that searches conducted in this tab are searched with word variations (except for the truncated terms), based on a Cochrane algorithm. In these instances, the Cochrane Library provides a note after the search string: (Word variations have been searched).
For example, if you search on “artificial intelligence” the algorithm will also include the phrase “artificially intelligent” to your search strategy. If you want to include these word variations in a complex search in Search Manager you must use the Search tab and then send the executed search to Search Manager to be included in your larger search strategy.
Search Manager will then format that search by enclosing it in parentheses and adding the field codes that were selected in the search tab (ex. ti,ab,kw) followed by the note (Word variations have been searched).
NOTE: Since word variations are ONLY added when the search is initiated from the Search tab, results will differ if the same search strategy is run in the Search tab as opposed to Search Manager. See example below to understand how results can differ in different circumstances:
Start from Search Tab
1. Enter terms and run search:
breast cancer AND hormone therapy
Results: 7,643 (split between Cochrane Reviews: 45 and Trials: 7,596)
2. Enter terms and send to search manager:
breast cancer AND hormone therapy
What is returned:
(breast cancer AND hormone therapy):ti,ab,kw
(Word variations have been searched)
Results: 7,643
----------------------------------------------------------------------
Start from Search Manager
1. Enter terms and run from Search Manager:
breast cancer AND hormone therapy
Results: 5,772
2. Enter terms and run from Search Manager:
(breast cancer AND hormone therapy):ti,ab,kw
Results: 5,412
Search Manager
The Search Manager is located within the Advanced Search tabs. Search Manager allows users to create and save complex multi-line searches. It supports the use of MeSH terms, logical operators, field labels, nesting, and wildcards. It also allows for unlimited search lines, and results can be viewed by search line (helpful to determine where issues might be in a strategy).
The search manager also allows for combining search lines by using their number (e.g. #1 AND #2; #1 OR #2) instead of the full content of each line, which can save time and space. It also can make it easier to see where and how concepts are being combined when developing complex searches.
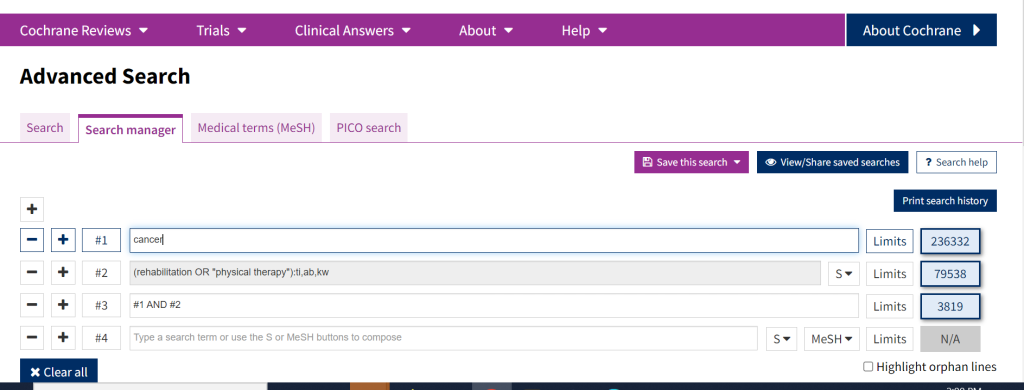
Searches conducted in the search tab can be sent to search manager, The search manager is necessary for conducting and saving complex searches, especially those for large literature reviews and systematic reviews. The search manager is the only type of search in Cochrane Library that allows capturing your search history – for that you need to click Print search history icon.
In order to save searches you have to be signed in your account from within your institutionally subscribed to Cochrane Library (Sign in is located in the right upper corner of the Search Manager Screen). Registering for an account allows you to set up email alerts based on your searches.
The Medical terms (MeSH) tab can also be used to search for relevant MeSH terms to add to your search, and then sent to the search manager.
PICO Search
The last advanced search tab is the PICO search which allows users to build a search according to the PICO framework (population, intervention, comparison, outcome). When using the PICO search as you type in search terms it will identify the specific “PICO vocabulary” that is relevant. This is necessary when using the PICO search as it will direct the terms to the specific PICO concept.
Other Search Functionalities in Cochrane
Boolean Operators
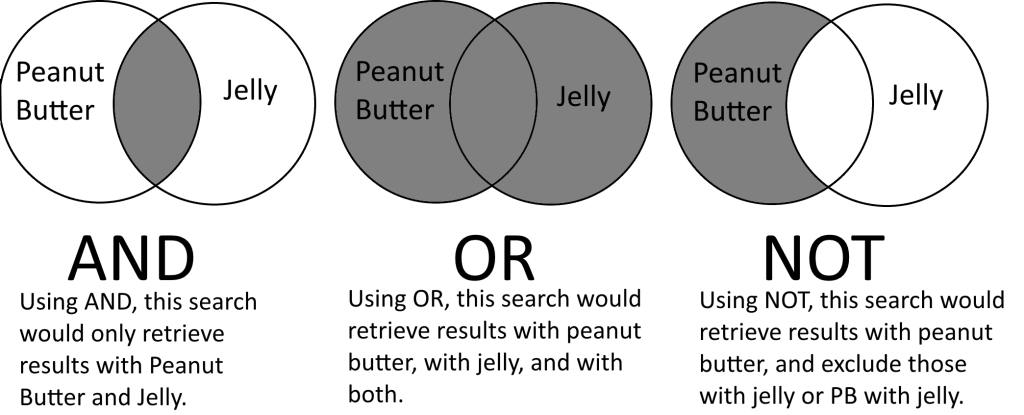
As in most online databases, Cochrane Library allows for users to search and combine terms using the Boolean Operators: AND, OR and NOT.
Exact Phrases
If you are searching for exact phrases, they should be enclosed in double quotations.
Proximity Operators
Cochrane Library allows for the use of Proximity operators, including: NEAR, NEAR/X and NEXT. Finds terms within close proximity to each other in the article or field(s) being searched.
NEAR
Finds the terms when they are within 6 words of each other. Terms can appear in either order.
cancer NEAR lung
Finds "lung cancer" and "cancer in the patient's lung"
NEAR/X
Finds the terms when they are within X words of each other where X = the maximum number of words between search terms. Terms can appear in either order.
cancer NEAR/3 lung
Finds "lung cancer" and "cancer of the lung", but NOT "cancer in the patient's lung"
NEXT
Finds the terms when they appear next to each other. Terms must appear in the order specified. Use the NEXT operator for phrase searching with wildcards.
cancer NEXT lung
Finds "lung cancer" but not "cancer of the lung" or "cancer in the patient's lung"
Truncation & Wildcards
Truncating a word means using a wildcard (*} which allows retrieving results with various ending of the word after truncation.
Do not use double quotes with truncation (*) anywhere in Cochrane Library searches, as truncation is ignored by Cochrane Library if used within double quotes. To overcome this, use the NEXT operator to search for an exact phrase with truncation.
Ex: instead of "heart transplant*" use (heart NEXT transplant*)
If you are interested in learning more about how to best use and search the Cochrane Library collection of databases, take a look at their e-books How to Search in Cochrane Library or Cochrane Library User Guide.