You need the National Center for Biomedical Ontology (NCBO) if you cannot complete one or both of the following tasks.
#1) define biomedical ontology.
#2) give an example of a biomedical ontology. To understand a biomedical ontology, you must first have basic knowledge of what an ontology is. The NCBO defines an ontology as “a kind of controlled vocabulary of well defined terms with specified relationships between those terms, capable of interpretation by both humans and computers.” Biomedical-specific ontologies are being developed to express and link data that are readable by computers and shared between data like experiments, articles, and gene expressions.
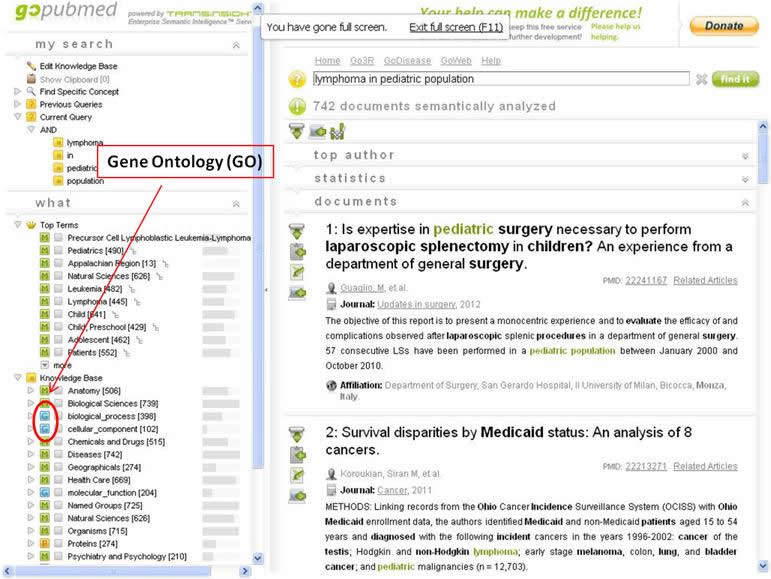
An example of a working biomedical ontology can be found in the GoPubMed database. In the left column you will see a list of terms preceded by either a green M or a blue G. The blue G denotes GoPubMed’s Gene Ontology (GO). This is their proprietary ontology of gene information that is linked to key terms when a user conducts a search. For example, when searching “lymphoma in pediatric population” you will notice three (3) GO terms in the left column. You are able to add any of these to your search builder to retrieve gene-specific information about this research topic. Notice how this GO works with your keyword search by mapping term concepts behind the scenes, making it easier for you to retrieve more valuable results. See the image below for a visual of the above example.